
The fungal glycoside product sordarin can specifically inhibit the activity of elongation factor EF-2 in fungal protein synthesis, but has no effect on human EF-2, making it a widely studied antifungal candidate drug. However, the structural complexity of sordarin poses a challenge for chemical total synthesis, which is faced with high input and low output. A comprehensive analysis of the biosynthetic pathway of sordarin can provide a solid foundation for the efficient and inexpensive production of sordarin by synthetic biology.
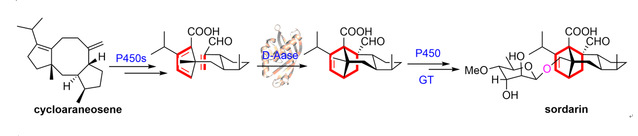
Recently, the research team led by Professor Huiming Ge and Professor Renxiang Tan at our university has fully elucidated the biosynthetic pathway from diterpene cycloaraneosene to sordarin, including the formation of the key Diels-Alder (D-A) reaction precursor of the 5/8/5 tricyclic skeleton under the catalysis of multiple P450s, followed by the formation of the dehydroabietene skeleton through the catalysis of a novel D-A enzyme SdnG. Further oxidation and glycosylation lead to the final product, and the catalytic mechanism of SdnG was inferred through the analysis of its crystal structure. The relevant achievements were recently published in Angew. Chem. Int. Ed. and selected as Hot Paper by the editor.
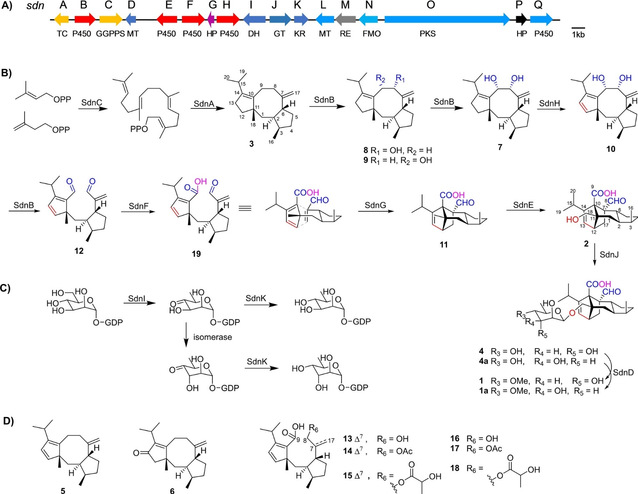
Figure 1 depicts the biosynthetic gene cluster (BGC) of Sordarin, as well as the intermediates and the proposed biosynthetic pathway.
Through preliminary research on sordarin and the use of bioinformatics tools, the sordarin biosynthetic gene cluster (BGC-sdn) was confirmed in the fungus Sordaria araneosa. Through in vivo gene knockout, heterologous expression, and identification of intermediate metabolites, the functions of several proteins were confirmed (Figure 1): SdnC and SdnA are responsible for the formation of the diterpene skeleton; SdnK, SdnJ, and SdnD are responsible for the synthesis of the sugar units; four P450 oxidases, SdnB, SdnE, SdnF, and SdnH, play important roles in the rearrangement of the molecular skeleton, with SdnB catalyzing the consecutive hydroxylation of C8 and C9 to form 7, and then breaking the C-C bond in the adjacent diol to form the dialdehyde product 12; SdnH catalyzes the desaturation of 7 to create the cyclopentadiene structure in the D-A reaction; SdnF converts the C9 aldehyde group in 12 to a carboxyl group, accelerating the generation of the D-A reaction product 11 within the molecule; and SdnE catalyzes a step of hydroxylation to form the sordaricin product. Thus, the complete biosynthetic pathway of sordarin has been elucidated. However, a conserved gene sdnG containing a SnoaL_2 domain was identified through comparative analysis of homologous gene clusters, suggesting the potential role of SdnG in the intramolecular D-A reaction, as evidenced by the identification of six open-ring products similar to the D-A reaction substrates in the sdnG mutant strain (13-18) (Figure 1).
To answer whether there is a D-A enzyme involved in the sordarin biosynthetic pathway, the authors purified the soluble protein of SdnG in vitro and synthesized compounds 12 and 19 for a series of enzymatic reaction experiments. The results (Figure 2) showed that compound 12, which contains a double aldehyde group, is not a substrate for SdnG, while compound 19 can undergo D-A reaction slowly and spontaneously, but can be quickly and completely catalyzed by SdnG to form the product 11 with dihydroisopyran skeleton. Therefore, the authors used in vitro enzymology experiments to demonstrate that the intramolecular D-A reaction in the sordarin biosynthetic process is a one-step enzymatic process.
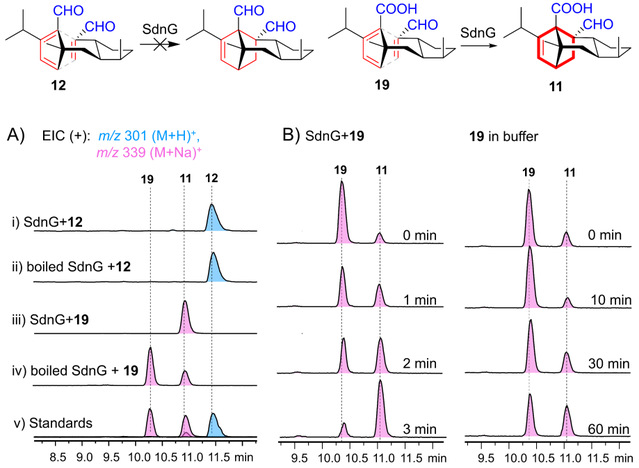
Figure 2. In vitro functional characterization of the D-A enzyme SdnG.
To further understand the catalytic mechanism of SdnG, the crystal structure of SdnG was obtained at a resolution of 2.70 Å (Figure 3A). Structural analysis revealed that its three-dimensional structure is similar to that of NgnD (Figure 3B), but with an active center located in a different position. Subsequently, docking of product 11 with the SdnG crystal structure (Figure 3G) showed that compound 11 is mainly surrounded by some hydrophobic residues, and the carboxyl group interacts with K127 and H72 through salt bridges, and with S91 through hydrogen bonds, while the aldehyde group is locked by R129 and Y41 through hydrogen bonds, promoting the D-A reaction. Subsequently, site-directed mutagenesis experiments demonstrated that these amino acids are closely related to the catalytic activity of SdnG (Figure 3H).

To further understand the catalytic mechanism of SdnG, the crystal structure of SdnG with a resolution of 2.70 Å was obtained (Fig. 3A). Structural analysis showed that its three-dimensional structure is similar to NgnD (Fig. 3B), but the active center is located in a different position. Subsequently, docking of product 11 with the SdnG crystal structure (Fig. 3G) showed that compound 11 is mainly surrounded by hydrophobic residues, while the carboxyl group interacts with K127 and H72 through salt bridges and with S91 through hydrogen bonds. The aldehyde group is locked by R129 and Y41 through hydrogen bonds, which promotes the occurrence of the D-A reaction. Subsequently, point mutation experiments proved that these amino acids are closely related to the catalytic activity of SdnG (Fig. 3H).
In summary, this work comprehensively elucidates the entire process of sordarin biosynthesis through in vivo gene knockout, intermediate structure identification, biotransformation, heterologous expression, and in vitro enzymatic experiments. It confirms the important functions of four P450 oxidases, SdnB, SdnE, SdnF, and SdnH, in the rearrangement process. It is worth noting that a novel D-A aldolase, SdnG, responsible for catalyzing the generation of the norbornene skeleton, was identified by the previous research team, further expanding the category of enzymatic D-A reactions in nature. This work also lays a solid foundation for further combinatorial biosynthesis research and microbial-based engineering production of the antifungal compound sordarin.
Dr. Shuang He Liu from the School of Life Sciences, Nanjing University, is the first author of the article, and Professors Hui Ming Ge, Ren Xiang Tan, and Associate Professor Bo Zhang from the School of Life Sciences, Nanjing University, are the corresponding authors. This research was supported by the National Key Research and Development Program, National Natural Science Foundation of China, Outstanding Youth Foundation, Youth Foundation, and the Fundamental Research Funds for the Central Universities.
Original link: https://onlinelibrary.wiley.com/doi/10.1002/anie.202205577
Article title: Biosynthesis of Sordarin Revealing a Diels-Alderase for the Formation of the Norbornene Skeleton.
