While mutation rates vary within genomes, suggestions that more selectively important DNA has a lower mutation rate are contentious not least because unbiased estimation of the mutation rate is challenging. A study by Monroe et al. (https://www.nature.com/articles/s41586-021-04269-6) also report in Arabidopsis that more important sequences have lower mutation rates and, while overlooking similar claims, suggest that this challenges “a long-standing paradigm regarding the randomness of mutation”.
Prof. Sihai Yang and Long Wang, from State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, together with Prof. Laurence D. Hurst and Alexander T. Ho from The Milner Centre for Evolution, Department of Biology and Biochemistry, University of Bath, find that in Monroe et al.’s work their mutation calling has abundant sequencing and analysis artefacts explaining why their data are not congruent with well-evidenced mutational profiles (Figure 1). The NJU and Bath research team has shown the key trends of Monroe et al’s work associated with sequence importance are consistent with well-described mutation-calling artefacts and are not resilient to reanalysis employing the higher quality components of their data (Figure 1). Although the NJU and Bath research team do not doubt that epigenetic marks such as methylation can affect mutation, given the above reasons and the failure to capture well described methyl C instability, the correlations evidenced by Monroe et al. between various marks and mutation rate variation should be treated with the same caution as their claim that mutation is rarer in more important sequences.
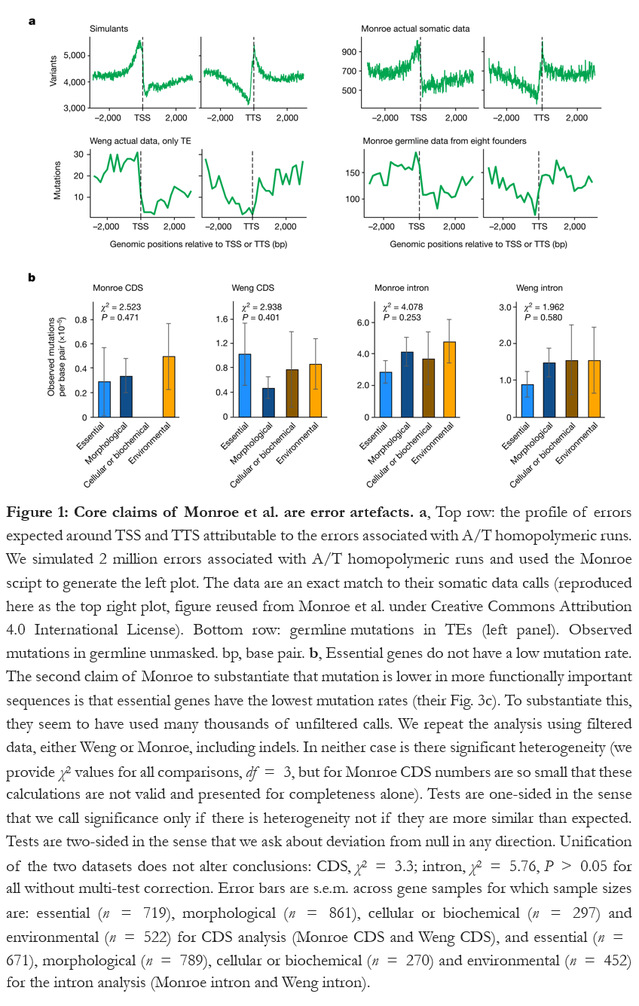
This “Matters arising” paper, published online on 26 July, 2023 in the journal Nature, is available at https://www.nature.com/articles/s41586-023-06314-y.
This work was supported by grants from the National Natural Science Foundation of China (grant numbers 31970236, 32270664, and 32170327).
